Analysis of Multiplexed, Single-Cell Clytia Medusae Experiments¶
Notebooks for reproducing all figures and analysis in the Whole Animal Multiplexed Single-Cell RNA-Seq Reveals Plasticity of Clytia Medusae Cell Types preprint. doi
Getting Started¶
All notebooks, saved as .ipynb's, can be run from Google Colab. Colab links are included in every notebook, just click on the symbol.
An intro to using Colab can be found here. Briefly, run each code cell by selecting the cell and executing Command/Ctrl+Enter. Code cells can be edited by simply clicking on the cell to start typing.
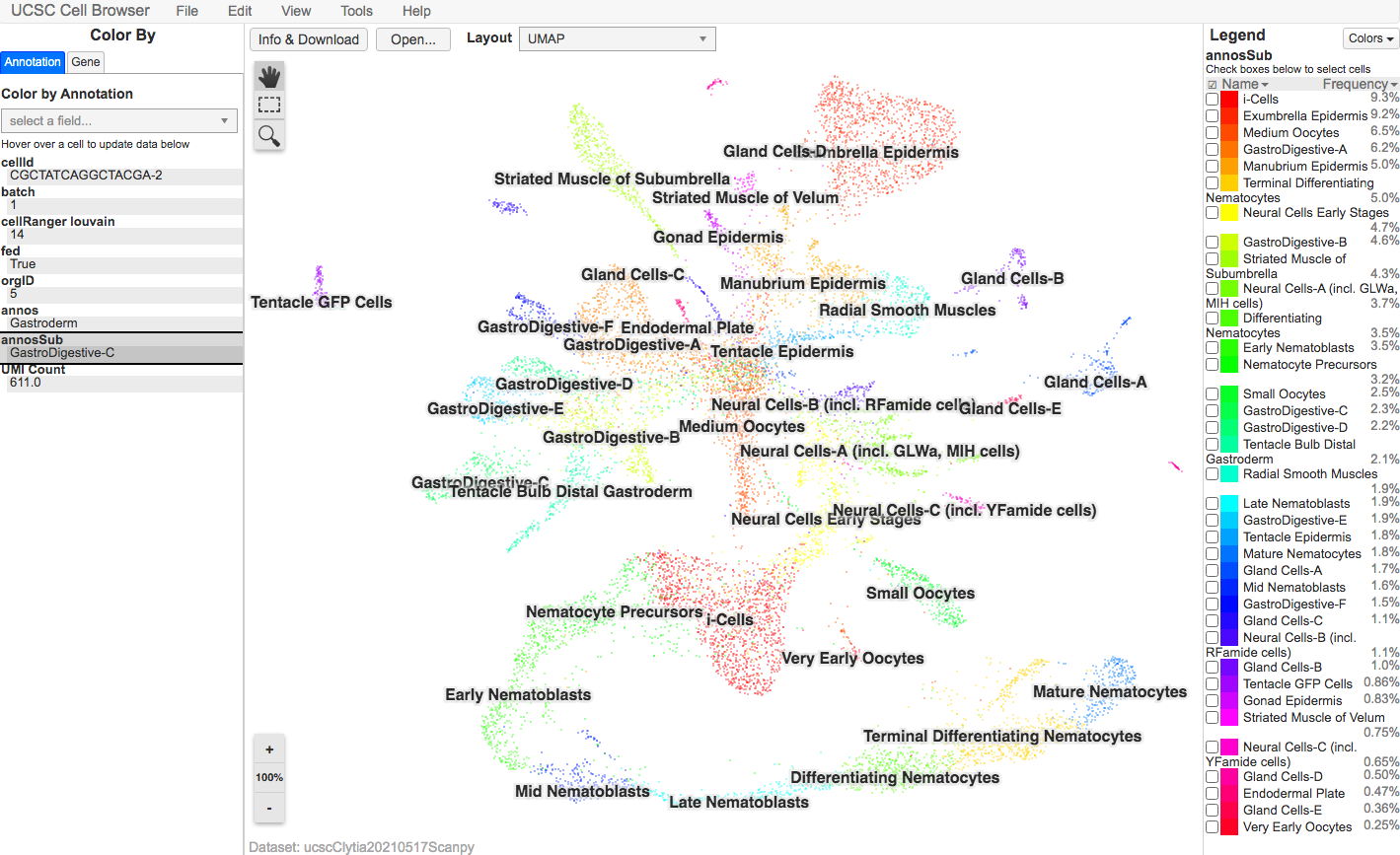
To view expression profiles of genes of interest use the UCSC Genome Browser Page with Interactive Browser or the Colab Notebook for generating images.
All saved/processed data used for analysis is streamed to the notebooks from CaltechData.
Quick Links¶
Data Files:
Raw Counts for Starved/Control Animals
Log-normalized Counts with HVGs for Starved/Control Animals
Interactive Browser:
UCSC Genome Browser Page with Interactive Browser
Analysis Notebooks¶
1) Pre-processing - All notebooks for ClickTag and cDNA Library Demultiplexing
a) ClickTag Pre-processing
- cellRangerClickTagCounts.ipynb
- Demultiplexing of ClickTag fastqs for starvation experiment from Cell Ranger bam files
- Input: Cell Ranger bam files of ClickTags
- Output: Count matrices for ClickTags
- kallistobusStarvClickTags.ipynb
- Demultiplexing of ClickTag fastqs for starvation experiment with kallisto-bustools workflow
- Input: Raw ClickTag fastqs
- Output: Count matrices for ClickTags (compared to previour count matrices)
- kallistobusStimClickTags.ipynb
- Demultiplexing of ClickTag fastqs for stimulation experiment with kallisto-bustools workflow
- Input: Raw ClickTag fastqs
- Output: Count matrices for ClickTags and filtered cell barcodes
b) cDNA Pre-processing
- filterStarvCells_ClickTags.ipynb
- Filtering of cell barcodes from cellRangerClickTagCounts ClickTag counts
- Input: ClickTag count matrices
- Output: Filtered cell barcodes and condition labels
- kallistoBusRuns_StarvAndStim.ipynb
- (Gene) Quantification of cDNA fastqs from starvation and stimulation experiments with kallisto-bustools
- Input: Raw cDNA fastqs
- Output: Anndata objects of cell x gene count matrices for each experiment
2) Cell Atlas and Perturbation Response - All notebooks for Clustering and Perturbation Response Analysis for Starvation Experiment
- cellRangerClustering_Starvation.ipynb
- Initial clustering of cells from starvation experiment from raw Cell Ranger (cDNA) matrices
- Input: Cell Ranger matrix
- Output: Anndata object of cell x gene count matrices for starvation experiment with clusters and highly variable genes
- starvation_Analysis.ipynb
- Clustering of cells from starvation experiment from kallisto-processed matrices (compared to cellRangerClustering_Starvation output)
- Input: kallisto anndata output
- Output: Anndata object of cell x gene count matrices for starvation experiment with clusters and highly variable genes and analysis of perturbation response
- deSeq2Analysis_StarvationResponse.ipynb
- DeSeq2 analysis for extracting perturbed genes from starved cells
- Input: Clustered anndata object from starvation_Analysis
- Output: Genes that are differentially expressed under starvation ('perturbed' genes)
- neuronSubpop_Analysis.ipynb
- Sub-clustering of neural cell types
- Input: Clustered anndata object from starvation_Analysis
- Output: Anndata object of cell x gene count matrices for neural cells with clusters and marker genes for subpopulations
- fullTrajAnalysis.ipynb
- Pseudotime analysis of neural, nematocyte, and gland cell types from the i-cell population
- Input: Clustered anndata object from starvation_Analysis
- Output: Ranking of genes contributing to pseudotime trajectories of neural and nematocyte cell types
3) Stimulation Data Analysis - Clustering and Perturbation Response Analysis for Stimulation Experiment
- stimulation_Analysis.ipynb
- Clustering of cells from stimulation experiment from kallisto-processed matrices
- Input: kallisto anndata output
- Output: Anndata object of cell x gene count matrices for stimulation experiment with clusters and highly variable genes and analysis of perturbation response
- deSeq2Analysis_StimulationResponse.ipynb
- DeSeq2 analysis for extracting perturbed genes from stimulated cells
- Input: Clustered anndata object from stimulation_Analysis
- Output: Genes that are differentially expressed under stimulation (DI/KCl) ('perturbed' genes)
4) Inter-Experimental Analysis - All Distance-based Analysis for Cell Type/State Delineation and Cross-Experiment Batch Effects
- allDistanceCalculations.ipynb
- Calculations for inter- and intra- cluster distances in starvation and stimulation experiments
- Input: Clustered anndata objects from stimulation_Analysis and starvation_Analysis
- Output: Merged dataset from both experiments and distance-based analysis
---------- For (User) Gene Searching and Plotting ----------
5) Explore Multiplexed Datasets - Notebooks for Exploring and Searching Cell Atlas
- MARIMBAAtlasSearchAndPlot.ipynb
- Plotting of genes of interest on MARIMBA-quantified anndata object
- Input: Clustered Anndata object from MARIMBAAnnosAnalysis, genes of interest
- Output: Gene expression profiles on cell atlas
- cellAtlasSearchAndPlot.ipynb
- Plotting of genes of interest on Trinity-quantified anndata object (used for all non-Marimba notebooks/main paper analysis)
- Input: Clustered anndata object from stimulation_Analysis, genes of interest
- Output: Gene expression profiles on cell atlas and neuron subpopulations
Authors¶
- Tara Chari
Acknowledgments¶
Several of the pre-processing workflows and initial analyses for the project were first implemented by Jase Gehring.