The goal of concordexR is to identify spatial homogeneous regions (SHRs) as defined in the recent manuscript, “Identification of spatial homogenous regions in tissues with concordex”. Briefly, SHRs are are domains that are homogeneous with respect to cell type composition. concordex relies on the the k-nearest-neighbor (kNN) graph to representing similarities between cells and uses common clustering algorithms to identify SHRs.
Installation
Versions of the concordexR package that do not enable clustering spatial data into spatial homogeneous regions (SHRs) are available for Bioconductor versions 3.17-19. The most recent version of the package is slated to be released on Bioconductor version 3.20.
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install(version="3.20")
#> 'getOption("repos")' replaces Bioconductor standard repositories, see
#> 'help("repositories", package = "BiocManager")' for details.
#> Replacement repositories:
#> CRAN: https://cran.rstudio.com/
#> Bioconductor version 3.20 (BiocManager 1.30.25), R 4.4.2 (2024-10-31)
#> Old packages: 'anytime', 'arrow', 'assorthead', 'BH', 'BiocCheck',
#> 'BiocNeighbors', 'Biostrings', 'bit', 'bit64', 'bookdown', 'class',
#> 'classInt', 'clue', 'cluster', 'cpp11', 'data.table', 'DelayedMatrixStats',
#> 'edgeR', 'evaluate', 'fitdistrplus', 'fontawesome', 'foreign',
#> 'GenomeInfoDb', 'geometry', 'ggmapinset', 'graph', 'graphlayouts', 'igraph',
#> 'IRanges', 'KernSmooth', 'knitr', 'later', 'limma', 'lme4', 'lpSolve',
#> 'lubridate', 'maps', 'MASS', 'MatrixGenerics', 'matrixStats', 'nnet',
#> 'parallelly', 'pillar', 'pkgbuild', 'processx', 'progressr', 'promises',
#> 'quantreg', 'Rcpp', 'RcppArmadillo', 'Rdpack', 'reticulate', 'Rfast',
#> 'rhdf5', 'rpart', 'RSQLite', 'Seurat', 'SFEData', 'shiny',
#> 'SingleCellExperiment', 'spatial', 'SpatialFeatureExperiment', 'spatialLIBD',
#> 'spatstat.data', 'spatstat.explore', 'spatstat.geom', 'spatstat.utils',
#> 'spData', 'spdep', 'stringdist', 'survival', 'systemfonts', 'terra',
#> 'testthat', 'textshaping', 'tidyterra', 'waldo', 'xfun', 'XML'
BiocManager::install("concordexR")
#> 'getOption("repos")' replaces Bioconductor standard repositories, see
#> 'help("repositories", package = "BiocManager")' for details.
#> Replacement repositories:
#> CRAN: https://cran.rstudio.com/
#> Bioconductor version 3.20 (BiocManager 1.30.25), R 4.4.2 (2024-10-31)
#> Warning: package(s) not installed when version(s) same as or greater than current; use
#> `force = TRUE` to re-install: 'concordexR'
#> Old packages: 'anytime', 'arrow', 'assorthead', 'BH', 'BiocCheck',
#> 'BiocNeighbors', 'Biostrings', 'bit', 'bit64', 'bookdown', 'class',
#> 'classInt', 'clue', 'cluster', 'cpp11', 'data.table', 'DelayedMatrixStats',
#> 'edgeR', 'evaluate', 'fitdistrplus', 'fontawesome', 'foreign',
#> 'GenomeInfoDb', 'geometry', 'ggmapinset', 'graph', 'graphlayouts', 'igraph',
#> 'IRanges', 'KernSmooth', 'knitr', 'later', 'limma', 'lme4', 'lpSolve',
#> 'lubridate', 'maps', 'MASS', 'MatrixGenerics', 'matrixStats', 'nnet',
#> 'parallelly', 'pillar', 'pkgbuild', 'processx', 'progressr', 'promises',
#> 'quantreg', 'Rcpp', 'RcppArmadillo', 'Rdpack', 'reticulate', 'Rfast',
#> 'rhdf5', 'rpart', 'RSQLite', 'Seurat', 'SFEData', 'shiny',
#> 'SingleCellExperiment', 'spatial', 'SpatialFeatureExperiment', 'spatialLIBD',
#> 'spatstat.data', 'spatstat.explore', 'spatstat.geom', 'spatstat.utils',
#> 'spData', 'spdep', 'stringdist', 'survival', 'systemfonts', 'terra',
#> 'testthat', 'textshaping', 'tidyterra', 'waldo', 'xfun', 'XML'Example
This is a basic example using concordex:
library(SFEData)
library(Voyager)
#> Loading required package: SpatialFeatureExperiment
#>
#> Attaching package: 'SpatialFeatureExperiment'
#> The following object is masked from 'package:base':
#>
#> scale
library(scran)
#> Loading required package: SingleCellExperiment
#> Loading required package: SummarizedExperiment
#> Loading required package: MatrixGenerics
#> Loading required package: matrixStats
#>
#> Attaching package: 'MatrixGenerics'
#> The following objects are masked from 'package:matrixStats':
#>
#> colAlls, colAnyNAs, colAnys, colAvgsPerRowSet, colCollapse,
#> colCounts, colCummaxs, colCummins, colCumprods, colCumsums,
#> colDiffs, colIQRDiffs, colIQRs, colLogSumExps, colMadDiffs,
#> colMads, colMaxs, colMeans2, colMedians, colMins, colOrderStats,
#> colProds, colQuantiles, colRanges, colRanks, colSdDiffs, colSds,
#> colSums2, colTabulates, colVarDiffs, colVars, colWeightedMads,
#> colWeightedMeans, colWeightedMedians, colWeightedSds,
#> colWeightedVars, rowAlls, rowAnyNAs, rowAnys, rowAvgsPerColSet,
#> rowCollapse, rowCounts, rowCummaxs, rowCummins, rowCumprods,
#> rowCumsums, rowDiffs, rowIQRDiffs, rowIQRs, rowLogSumExps,
#> rowMadDiffs, rowMads, rowMaxs, rowMeans2, rowMedians, rowMins,
#> rowOrderStats, rowProds, rowQuantiles, rowRanges, rowRanks,
#> rowSdDiffs, rowSds, rowSums2, rowTabulates, rowVarDiffs, rowVars,
#> rowWeightedMads, rowWeightedMeans, rowWeightedMedians,
#> rowWeightedSds, rowWeightedVars
#> Loading required package: GenomicRanges
#> Loading required package: stats4
#> Loading required package: BiocGenerics
#>
#> Attaching package: 'BiocGenerics'
#> The following object is masked from 'package:SpatialFeatureExperiment':
#>
#> saveRDS
#> The following objects are masked from 'package:stats':
#>
#> IQR, mad, sd, var, xtabs
#> The following objects are masked from 'package:base':
#>
#> anyDuplicated, aperm, append, as.data.frame, basename, cbind,
#> colnames, dirname, do.call, duplicated, eval, evalq, Filter, Find,
#> get, grep, grepl, intersect, is.unsorted, lapply, Map, mapply,
#> match, mget, order, paste, pmax, pmax.int, pmin, pmin.int,
#> Position, rank, rbind, Reduce, rownames, sapply, saveRDS, setdiff,
#> table, tapply, union, unique, unsplit, which.max, which.min
#> Loading required package: S4Vectors
#>
#> Attaching package: 'S4Vectors'
#> The following object is masked from 'package:utils':
#>
#> findMatches
#> The following objects are masked from 'package:base':
#>
#> expand.grid, I, unname
#> Loading required package: IRanges
#> Loading required package: GenomeInfoDb
#> Loading required package: Biobase
#> Welcome to Bioconductor
#>
#> Vignettes contain introductory material; view with
#> 'browseVignettes()'. To cite Bioconductor, see
#> 'citation("Biobase")', and for packages 'citation("pkgname")'.
#>
#> Attaching package: 'Biobase'
#> The following object is masked from 'package:MatrixGenerics':
#>
#> rowMedians
#> The following objects are masked from 'package:matrixStats':
#>
#> anyMissing, rowMedians
#> Loading required package: scuttle
library(bluster)
library(concordexR)
sfe <- McKellarMuscleData("small")
#> see ?SFEData and browseVignettes('SFEData') for documentation
#> loading from cache
clusters <- quickCluster(sfe, min.size=2, d=15)
nbc <- calculateConcordex(
sfe,
clusters,
n_neighbors=10,
BLUSPARAM=KmeansParam(2)
)
colData(sfe)[["shr"]] <- attr(nbc, "shrs")
plotSpatialFeature(sfe, features="shr")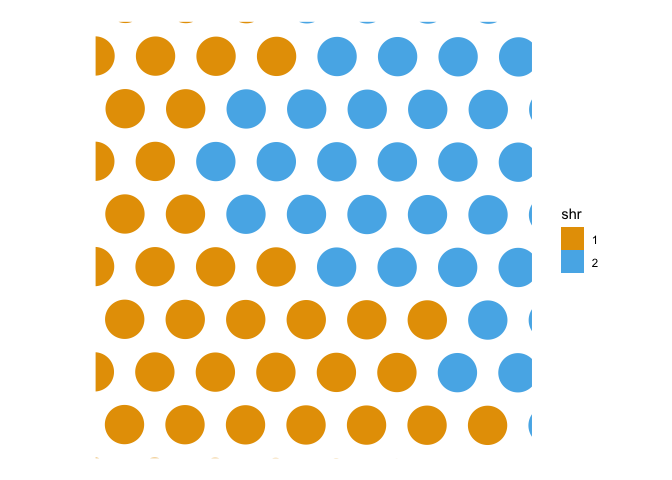
Citation
If you’d like to use the concordexR package in your research, please cite our recent bioRxiv preprint
Jackson, K.; Booeshaghi, A. S.; Gálvez-Merchán, Á.; Moses, L.; Chari, T.; Pachter, L. Quantitative assessment of single-cell RNA-seq clustering with CONCORDEX. bioRxiv (Cold Spring Harbor Laboratory) 2023. https://doi.org/10.1101/2023.06.28.546949.
@article {Jackson2023.06.28.546949, author = {Jackson, Kayla C. and Booeshaghi, A. Sina and G{’a}lvez-Merch{’a}n, {’A}ngel and Moses, Lambda and Chari, Tara and Kim, Alexandra and Pachter, Lior}, title = {Identification of spatial homogeneous regions in tissues with concordex}, year = {2024}, doi = {10.1101/2023.06.28.546949}, publisher = {Cold Spring Harbor Laboratory}, URL = {https://www.biorxiv.org/content/early/2024/07/18/2023.06.28.546949}, journal = {bioRxiv} }