This function plots the images in SFE objects without plotting geometries. When showing axes, the numbers are coordinates within the image itself and have the same units as the spatial extent, but are not the actual spatial extent when plotting multiple samples to avoid excessive empty space.
Usage
plotImage(
sfe,
sample_id = "all",
image_id = NULL,
channel = NULL,
ncol = NULL,
bbox = NULL,
maxcell = 5e+05,
show_axes = FALSE,
dark = FALSE,
palette = colorRampPalette(c("black", "white"))(255),
normalize_channels = FALSE
)Arguments
- sfe
A
SpatialFeatureExperimentobject.- sample_id
Sample(s) in the SFE object whose cells/spots to use. Can be "all" to compute metric for all samples; the metric is computed separately for each sample.
- image_id
ID of the image(s) to plot. If
NULL, then the first image present is plotted. Can be a vector of IDs to use different grayscale images for different channels. The vector can be named ('r', 'g', 'b'), to assign channels to images. The vector must be named if it's length 2.- channel
Numeric vector indicating which channels in a multi-channel image to plot. If
NULL, grayscale is plotted if there is 1 channel and RGB for the first 3 channels. The numeric vector can be named (r, g, b) to indicate which channel maps to which color. The RGB colorization may not be colorblind friendly. This argument cannot be specified whileimage_idis a named vector to plot different grayscale images as different channels.- ncol
Number of columns if plotting multiple features. Defaults to
NULL, which means using the same logic asfacet_wrap, which is used bypatchwork'swrap_plotsby default.- bbox
A bounding box to specify a smaller region to plot, useful when the dataset is large. Can be a named numeric vector with names "xmin", "xmax", "ymin", and "ymax", in any order. If plotting multiple samples, it should be a matrix with sample IDs as column names and "xmin", "ymin", "xmax", and "ymax" as row names. If multiple samples are plotted but
bboxis a vector rather than a matrix, then the same bounding box will be used for all samples. You may see points at the edge of the geometries if the intersection between the bounding box and a geometry happens to be a point there. IfNULL, then the entire tissue is plotted.- maxcell
Maximum number of pixels to plot in the image. If the image is larger, it will be resampled so it have less than this number of pixels to save memory and for faster plotting. We recommend reducing this number when plotting multiple facets.
- show_axes
Logical, whether to show axes.
- dark
Logical, whether to use dark theme. When using dark theme, the palette will have lighter color represent higher values as if glowing in the dark. This is intended for plotting gene expression on top of fluorescent images.
- palette
Vector of colors to use to color grayscale images.
- normalize_channels
Logical, whether to normalize each channel of the image individually. Should be
FALSEfor bright field color images such as H&E but should set toTRUEfor fluorescent images.
Examples
library(SFEData)
library(SpatialFeatureExperiment)
fn <- XeniumOutput("v2", file_path = "xenium_example")
#> see ?SFEData and browseVignettes('SFEData') for documentation
#> downloading 1 resources
#> retrieving 1 resource
#> loading from cache
#> The downloaded files are in /home/runner/work/voyager/voyager/docs/reference/xenium_example/xenium2
# Weird RBioFormats null pointer error the first time it's run
try(sfe <- readXenium(fn))
#> >>> Cell segmentations are found in `.parquet` file(s)
#> >>> Reading cell and nucleus segmentations
#> >>> Making MULTIPOLYGON nuclei geometries
#> >>> Making POLYGON cell geometries
#> Sanity checks on cell segmentation polygons:
#> >>> ..found 132 cells with (nested) polygon lists
#> >>> ..applying filtering
#> >>> Checking polygon validity
#> >>> Saving geometries to parquet files
#> >>> Reading cell metadata -> `cells.parquet`
#> >>> Reading h5 gene count matrix
#> >>> filtering cellSeg geometries to match 6272 cells with counts > 0
#> >>> filtering nucSeg geometries to match 6158 cells with counts > 0
sfe <- readXenium(fn)
#> >>> Preprocessed sf segmentations found
#> >>> Reading cell and nucleus segmentations
#> >>> Reading cell metadata -> `cells.parquet`
#> >>> Reading h5 gene count matrix
#> >>> filtering cellSeg geometries to match 6272 cells with counts > 0
#> >>> filtering nucSeg geometries to match 6158 cells with counts > 0
# Plot one channel
plotImage(sfe, image_id = "morphology_focus", channel = 1L)
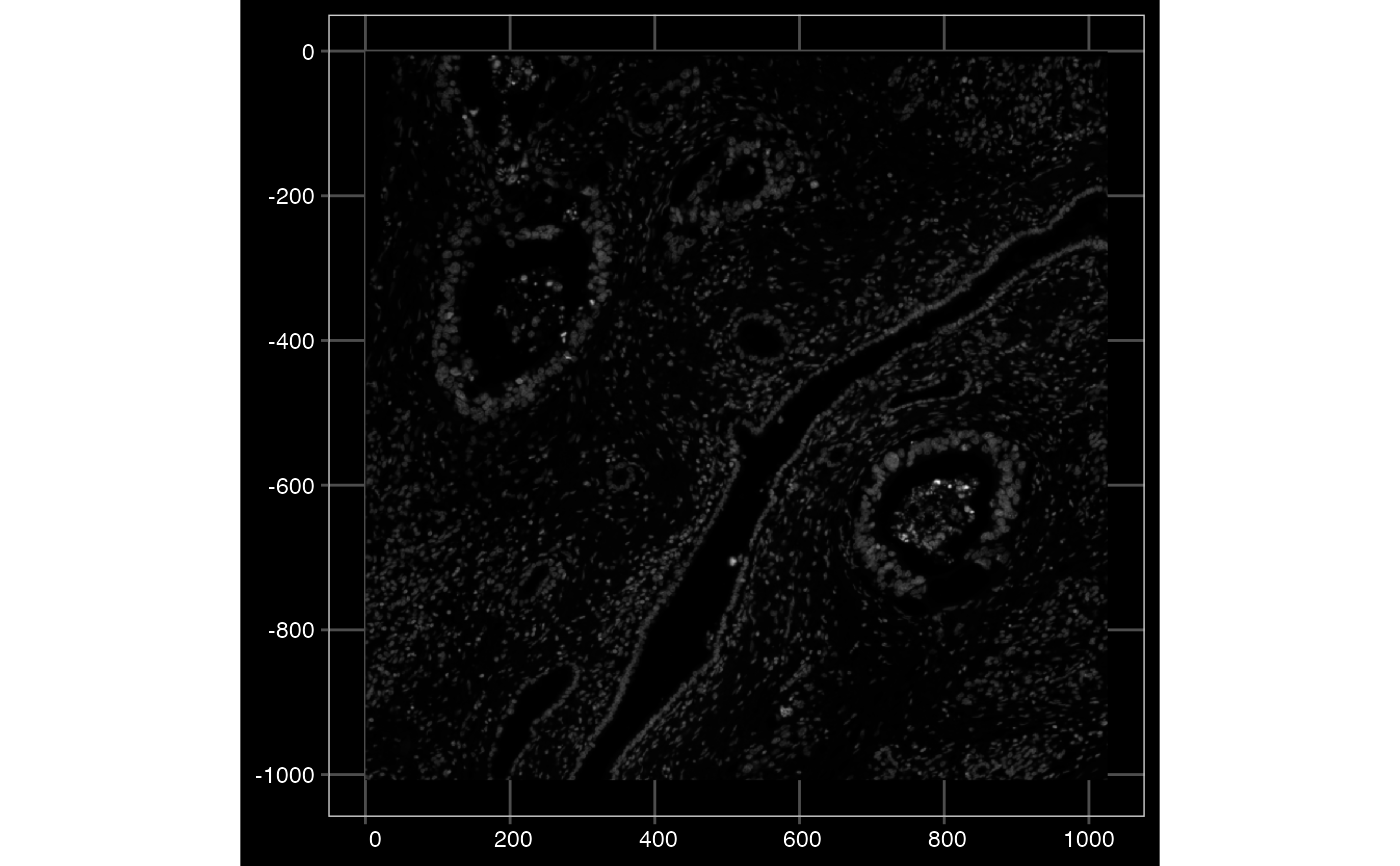
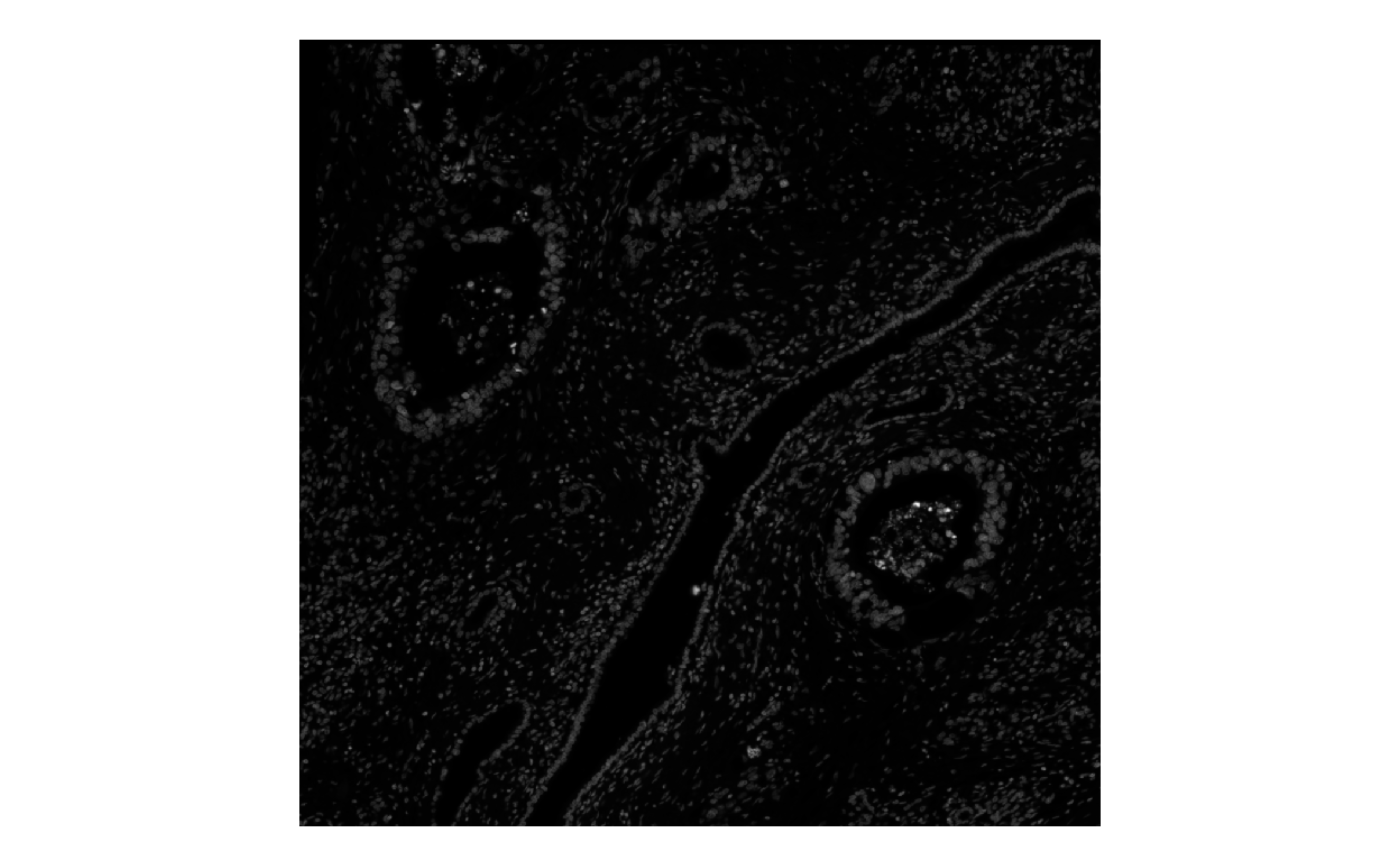 plotImage(sfe, image_id = "morphology_focus", channel = 1L, show_axes = TRUE, dark = TRUE)
plotImage(sfe, image_id = "morphology_focus", channel = 1L, show_axes = TRUE, dark = TRUE)
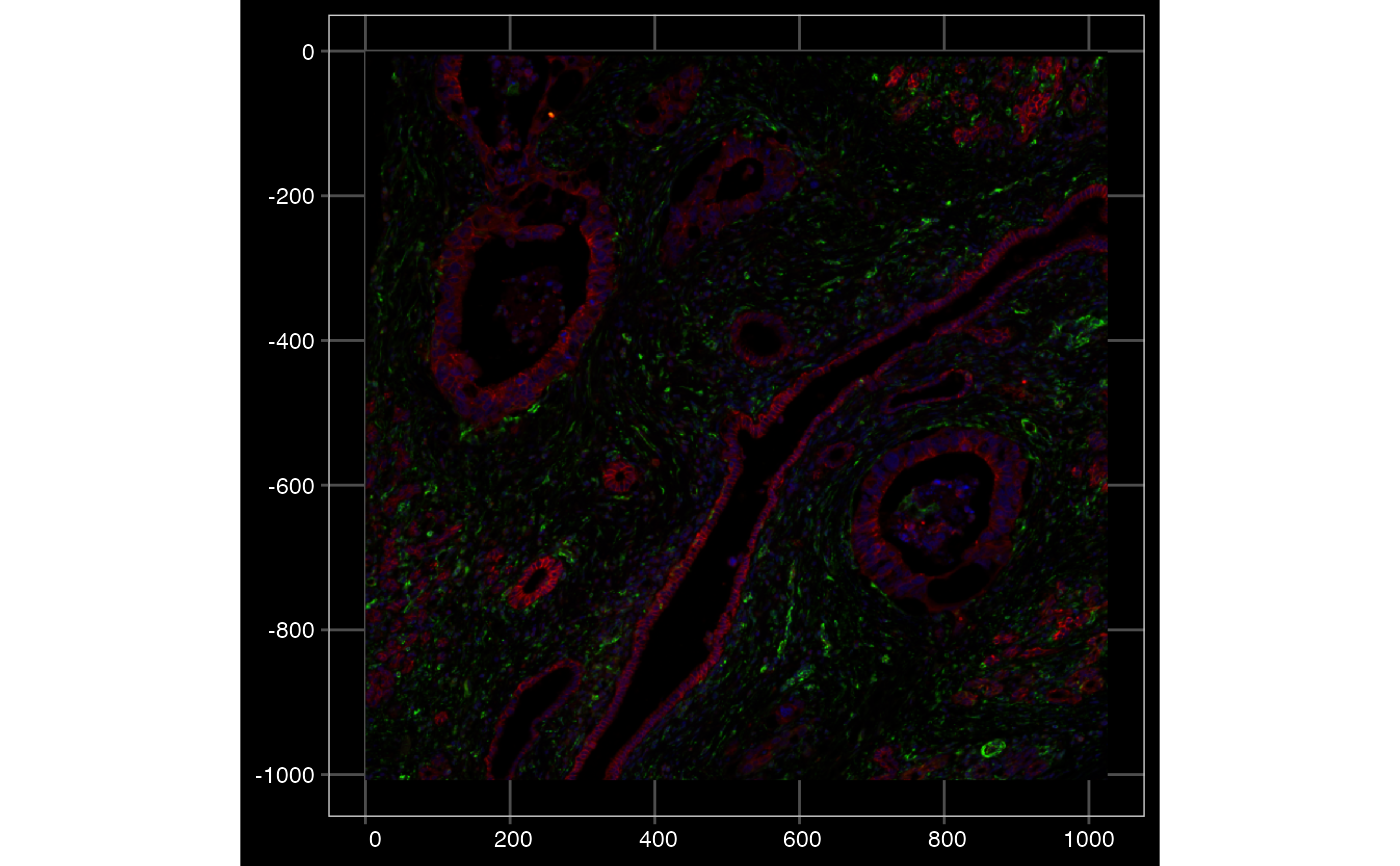
 # Colorize based on different channels
plotImage(sfe, image_id = "morphology_focus", channel = c(2,4,1), show_axes = TRUE, dark = TRUE)
# Colorize based on different channels
plotImage(sfe, image_id = "morphology_focus", channel = c(2,4,1), show_axes = TRUE, dark = TRUE)
 unlink("xenium_example", recursive = TRUE)
unlink("xenium_example", recursive = TRUE)