💡 Active users of this documentation website
Automatically updates every day at 16:00 UTC.
⬇️ Daily gget downloads

Automatically updates every Sunday at 23:55 UTC.
🧑🤝🧑 Dependent software
The following applications build on gget:
- gget-mcp
"MCP server providing a powerful bioinformatics toolkit for genomics queries and analysis, wrapping the popular
ggetlibrary."- Other tools using this or other
ggetMCP servers:
- Other tools using this or other
- Biomni
A General-Purpose Biomedical AI Scientist being developed at Stanford and Genentech. - PerTurboAgent
A Self-Planning Agent for Boosting Sequential Perturb-seq Experiments."We [...] use packages gget and blitzgsea for data enrichment analysis"
- Scientific skills for Claude by K-Dense-AI
" This repository contains 138 scientific skills organized across multiple domains. Each skill provides comprehensive documentation, code examples, and best practices for working with scientific libraries, databases, and tools.
🧬 Bioinformatics & Genomics
Genomic tools: gget, ..." - Therapeutics Data Commons (TDC)
Artificial intelligence foundation for therapeutic science (source code, Nat Chem Bio paper) by Harvard's Artificial Intelligence for Medicine and Science lab. - BioDiscoveryAgent
BioDiscoveryAgent is an LLM-based AI agent for closed-loop design of genetic perturbation experiments (preprint) by the Stanford Network Analysis Project. - DeepChopper
Language models to identify chimeric artificial reads in NanoPore direct-RNA sequencing data by the Yang lab at Northwestern. - BRAD
A LLM powered chatbot for bioinformatics (documentation, project main page). - scPRINT
scPRINT is a large transformer model built for the inference of gene networks (connections between genes explaining the cell's expression profile) from scRNAseq data (preprint). - AnoPrimer
AnoPrimer is a Python package for primer design in An. gambiae and An. funestus, whilst considering genetic variation in wild whole-genome sequenced specimens in malariagen_data. - AvaTaR
Optimizing LLM Agents for Tool-Assisted Knowledge Retrieval (NeurIPS 2024) by James Zou Lab at Stanford University. - GRLDrugProp
Graph representation learning for modelling drug properties. - MicrobioLink2
A computational tool that analyzes the impact of host-microbe interaction on downstream signaling in human cells and tissues. - Rust implementation of gget: https://github.com/noamteyssier/ggetrs
- https://github.com/Superbio-ai/getbio
- https://github.com/yonniejon/AchillesPrediction
- https://github.com/ELELAB/cancermuts
- https://github.com/Benoitdw/SNPrimer
- https://github.com/louisjoecodes/a16z-hackathon-project
- https://github.com/EvX57/BACE1-Drug-Discovery
- https://github.com/vecerkovakaterina/hidden-genes-msc
- https://github.com/vecerkovakaterina/llm_bioinfo_agent
- https://github.com/greedjar74/upstage_AI_Lab
- https://github.com/alphavector/all
See more: https://github.com/pachterlab/gget/network/dependents
📃 Featured publications using gget
- David Bradley et al., The fitness cost of spurious phosphorylation. The EMBO Journal (2024). DOI: 10.1038/s44318-024-00200-7
"Phosphosites matching to putative short linear motifs (SLiMs) were extracted using the gget elm resource"
- Mikael Nilsson et al., Resolving thyroid lineage cell trajectories merging into a dual endocrine gland in mammals. Nature Portfolio (under review) (2024). DOI: 10.21203/rs.3.rs-5278325/v1
"Enrichment analysis of differentially expressed genes [...] was performed using gget enrichr"
- Avasthi P et al., Repeat expansions associated with human disease are present in diverse organisms. Arcadia (2024). DOI: 10.57844/arcadia-e367-8b55
"We used the gget package using the gget.blast command [...] to BLAST our proteins of interest against the non-redundant NCBI protein database"
- Ibrahim Al Rayyes et al., Single-Cell Transcriptomics Reveals the Molecular Logic Underlying Ca2+ Signaling Diversity in Human and Mouse Brain. bioRxiv (2024). DOI: 10.1101/2024.04.26.591400
"The gget package was used to perform the gene set enrichment analyses with databases Reactome_2022 and KEGG_2021_Mouse."
- Beatriz Beamud et al., Genetic determinants of host tropism in Klebsiella phages. Cell Reports (2023). DOI: 10.1016/j.celrep.2023.112048
"Proteins with seemingly enzymatic activity were confirmed to have the characteristic β-helical domain using gget AlphaFold2"
- Kimberly Siletti et al., Transcriptomic diversity of cell types across the adult human brain. Science (2023). DOI: 10.1126/science.add7046
"The gget package (https://github.com/pachterlab/gget) was used to perform all gene ontologyenrichment analysis"
- David R. Blair & Neil Risch. Dissecting the Reduced Penetrance of Putative Loss-of-Function Variants in Population-Scale Biobanks. medRxiv (2024). DOI: 10.1101/2024.09.23.24314008
"Additional gene and transcript information (exon-intron boundaries, 5' and 3' UTRs, full coding and amino acid sequences) was downloaded using gget"
- Shanmugampillai Jeyarajaguru Kabilan et al., Molecular modelling approaches for the identification of potent Sodium-Glucose Cotransporter 2 inhibitors from Boerhavia diffusa for the potential treatment of chronic kidney disease. Journal of Computer-Aided Molecular Design (under review) (2024). DOI: 10.21203/rs.3.rs-4520611/v1
- Joseph M Rich et al., The impact of package selection and versioning on single-cell RNA-seq analysis. bioRxiv (2024). DOI: 10.1101/2024.04.04.588111
- Sanjay C. Nagi et al., AnoPrimer: Primer Design in malaria vectors informed by range-wide genomic variation. Wellcome Open Research (2024).
- Yasmin Makki Mohialden et al., A survey of the most recent Python packages for use in biology. NeuroQuantology (2023). DOI: 10.48047/NQ.2023.21.2.NQ23029
- Nicola A. Kearns et al., Generation and molecular characterization of human pluripotent stem cell-derived pharyngeal foregut endoderm. Cell Reports (2023). DOI: 10.1016/j.devcel.2023.08.024
- Jonathan Rosenski et al., Predicting gene knockout effects from expression data. BMC Medical Genomics (2023). DOI: 10.1186/s12920-023-01446-6
- Peter Overby et al., Pharmacological or genetic inhibition of Scn9a protects beta-cells while reducing insulin secretion in type 1 diabetes. bioRxiv (2023). DOI: 10.1101/2023.06.11.544521
- Mingze Dong et al., Deep identifiable modeling of single-cell atlases enables zero-shot query of cellular states. bioRxiv (2023). DOI: 10.1101/2023.11.11.566161
See more: Google Scholar
📰 gget in the news
- Oreate AI blog post: "Exploring AlphaFold2: A Guide to GitHub Repositories and Colab Resources"
gget opentargetsrelease blog post by the Open Targets platform- Documentary short film about gget: https://youtu.be/cVR0k6Mt97o
- Podcast episode for the Prototype Fund Public Interest Podcast about the importance of open-source software and its role in academic research (in German): https://public-interest-podcast.podigee.io/33-pips4e4
- Prototype Fund announcement: https://prototypefund.de/project/gget-genomische-datenbanken
🚂 gget code repository traffic
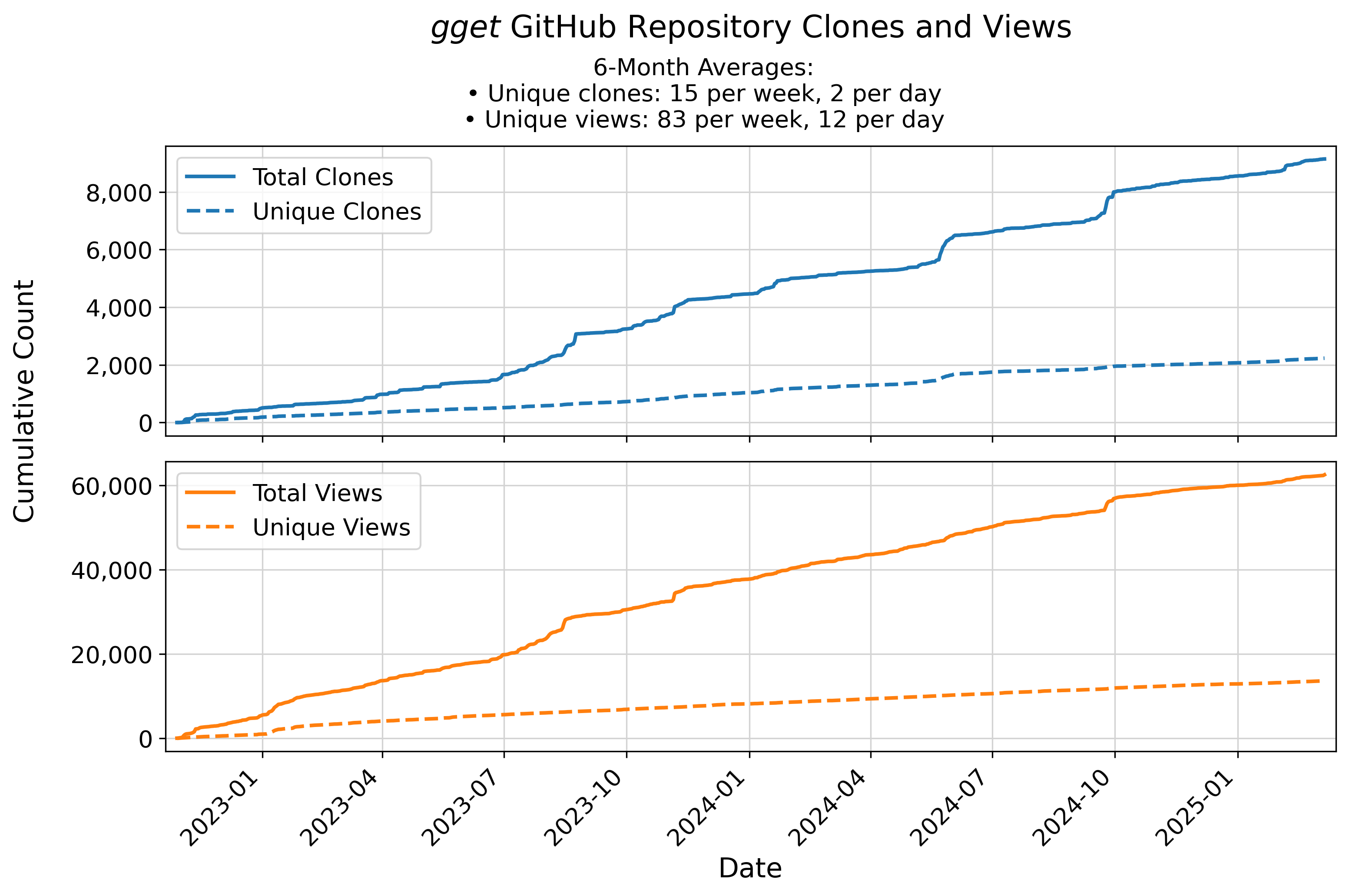 Updates automatically every week on Sunday at 23:55 (UTC).
Updates automatically every week on Sunday at 23:55 (UTC).
